Review Article - Journal of Apitherapy (2022)
Review on Honeybee as a Simple Model Animal for Microbiome Research Work
Muhammad Fahad Raza1*, Bilal Atta2, Muhammad Ammar Raza3 and Muhammad Asad42Department of Entomology, Rice Research Institute, Sheikhupura, Pakistan
3Department of Field Crops, Siirt University, Siirt, Turkey
4Department of Applied Ecology, Fujian Agriculture and Forestry University, Fuzhou, China
Muhammad Fahad Raza, Department of Entomology, University of Agriculture, Faisalabad, Pakistan, Email: fbiuaf91@gmail.com
Received: 01-Mar-2022, Manuscript No. JAPITHERAPY-22-60418; Editor assigned: 03-Mar-2022, Pre QC No. JAPITHERAPY-22-60418 (PQ); Reviewed: 18-Mar-2022, QC No. JAPITHERAPY-22-60418; Revised: 23-Mar-2022, Manuscript No. JAPITHERAPY-22-60418 (R); Published: 30-Mar-2022
Abstract
Microbiome research of the gut is an emerging discipline that seeks to understand better the functional and ecological dynamics of microbiota. The gut microbiota of honeybees is a unique community to investigate, as honeybees are ecologically essential pollinators of several crops grown for human consumption and they produce valuable products like royal jelly, wax and honey. Most importantly, the gut environment of Apis mellifera has unique characteristics that make it an excellent model system. This review discusses the honeybee gut microbiota significance, its effect on behavior and endocrine signalling, neurological effect of gut microbiota, perturbation of native microbiota, and structural differences of gut microbiota in summer and winter. This review also outlined the microbiome research on the traditional biomedical model’s honeybee, zebrafish, Drosophila melanogaster, and Caenorhabditis elegans command outstanding research resources tools are addressed. This review highlights the honeybee as a promising model insect to better understand honeybee gut microbiota, facilitating microbiome research and bee microbiota in general, and supporting future prospective.
Keywords
Apis mellifera; gut microbiota; Drosophila melanogaster; Caenorhabditis elegans; Danio rerio
Introduction
The microbiomes study of animals has extensively appeared as a new discipline, possibly initiated by sequencing technologies to find the functional traits of microorganisms and taxonomic identity without cultivation. During the past 10-15 years, the rise of microbiomes science has provided a way to minimize chronic diseases of humans, particularly immunological, metabolic abnormality and anxiety health disorders. Recently, microbiome science is usually used as a biomedical discipline that emphasizes humans’ microbiology, helped by research on the laboratory mouse [1-3]. The most significant contribution to understanding the animal microbiomes has been provided by the research on the simple model animal such as lower vertebrates and invertebrates, which are associated with microbiomes with a lower level of taxonomic diversity. Relative to mammals, these simplistic systems provide simple protocols for manipulation of microbiota population and distribute role to specific copiotrophic microbes, develop cost-effective experiments within less period, allow the intricate experimental designs and unusually for invertebrates, avoid the critical animal welfare problems elevated by research on mammals [4,5].
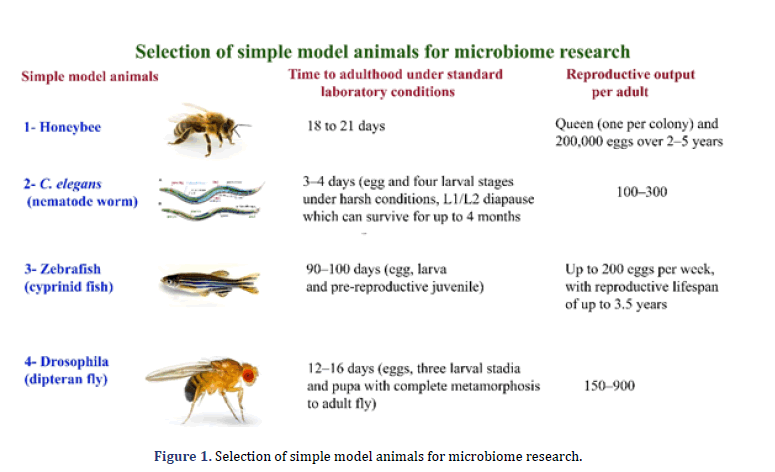
The research on the gut microbiome is unique and valuable for a better understanding of functional, biological, and ecological processes inside hosts. The gut microbiome performs many tasks with important significance for overall fitness and the host. Gut microbial communities play an essential role in many host organisms to break down complex carbohydrates to immunomodulation [6,7]. The primary and critical characteristics in symbiotic gut microorganisms are to expose uncharacterized mechanisms, in addition to the development of prophylactic and clinical handlings that help both animals and humans [8,9]. The fruit fly Drosophila melanogaster, honeybee Apis mellifera, nematode Caenorhabditis elegan, and the zebrafish Danio rerio are simple model organisms. These biomedical models are gaining more attention because microbiome research is a direct way to effectively research neurobiology and immune function at the molecular level using these systems and animal development. This review discusses an outline for microbiome research of simple model animals, including their importance and drawbacks. The review focuses on using simple model animals for microbiome research particularly (Figure 1).
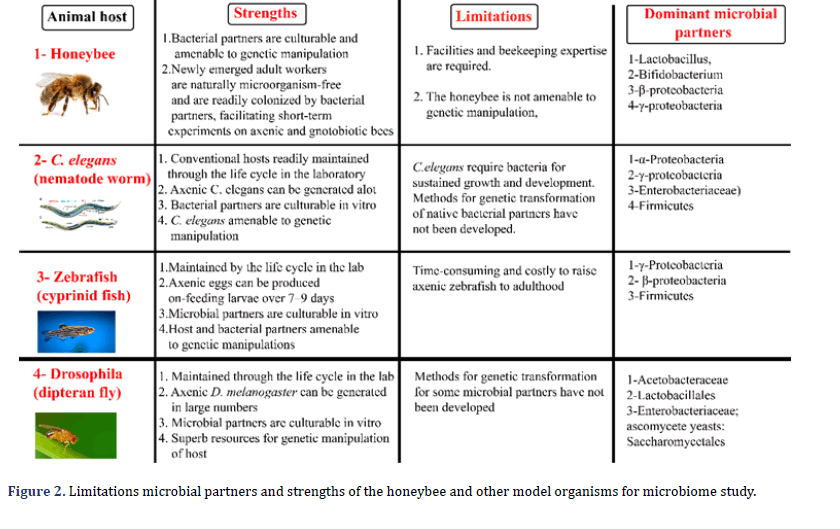
The A. mellifera, D. melanogaster, zebrafish and C. elegans have been primarily genetic models. The emerging and valuable model animals for microbiome research are A. mellifera due to microbiome interaction with xenobiotics, pesticides, and complicated. The A. mellifera has a strong technical and scientific source due to its significance for honey production and pollination. The zebrafish is an essential biomedical model in vertebrates, although its generation period is significantly more extended than C. elegans and D. Melanogaster (Figure 2). These traditional model animals are gradually being implemented for host-microbiome interactions as the parallels in processes and patterns across the animal kingdom [10, 11].
Outlines for Simple Model Animals
The most important and emerging field is gut microbiota research, which increases understanding of gut environments’ functional and ecological dynamics. The study of honeybee gut microbiota highly rewards the community because honeybees produce valued commodities such as honey and wax for human utilization, also playing a role as primary pollinators of many crops. The A. mellifera gut habitat is a valuable model system due to its unique characteristics [12,13]. The genetic manipulation tools and standardized laboratory protocols related to microbiome research for A. mellifera, D. melanogaster, and zebrafish and C. elegans are already developed to facilitate the research community. Surface¬ sterilizing eggs can generate large numbers of axenic hosts usually using bleach, and raise the animals in sterile tubes or dishes [14,15]. The desired microorganisms resulting from gut homogenates and fecal pellets are mixed with a culture medium to obtain gnotobiotic animals [16].
The D. melanogaster axenic can maintain through multiple generations on nutrient-rich media. However, several experiments related to axenic of zebrafish and C. elegans are limited to young larvae. The bacterial community requires for development and sustained growth C. elegans. Hence, this necessity can be fulfilled by a medium supplemented with artificial liposome nanoparticles [17,18].
The zebrafish axenic can be reared to the adult stage, but the process is expensive and laborious. The commercially available sterile fish food for axenic zebrafish is toxic, to produce and administer the live food such as a member of the genus Tetrahymena is time-consuming. These are the main problem related to feeding the axenic zebrafish [19,20]. All developmental stages of C. elegans and Zebrafish larvae have significance. Some critical consideration has been offered to the design and explanation of microbiota studies for C. elegans and D. melanogaster because their hosts are microbes and various microorganisms Racz digest the food items, which make an essential contribution to host nutrition [21,22]. However, zebrafish are omnivores, which feed on crustaceans, aquatic insects, and plant material. The C. elegans and D. melanogaster administer microorganisms for standard laboratory protocols. The most common laboratory diet for C. elegans and D. melanogaster is dead yeast [23-26]. Nevertheless, viable microorganisms can be separated regularly from the gut of both species, and some bacteria can persist and often proliferate for an extended period in the gut of D. melanogaster [27-29].
Significance of the honeybee gut microbiota
A. mellifera (the Western honeybee) is a pollinator safeguarding food security. A. mellifera is performing pollination activity for more than 92 major economic crops. Products of A. mellifera, such as wax and honey, contribute to the economic importance of honeybee, accompanying a billion-dollar pollination industry. The high demand for modern agricultural practices (such as agrochemical usage and pesticide), commercial pollination facilities, and environmental problems (such as pathogen spread and poor nutrition) have challenged colony sustainability [30- 32]. The study related to the gut microbiota of A. mellifera has developed rapidly. Recent studies have shown distinct characteristics, which make the unique gut environment of the A. mellifera amongst other insects. Honey bees have been studied as model insects of social behaviour, developmental biology [33]. and behavioral disorders. The economic value of honeybees, especially from pollination, is assessed in billions of dollars annually [34-37]. The worldwide concern about increasing seasonal mortality rates of beehives motivates the research regarding ecological factors affecting bee health, including toxins, nutrition pathogens, and parasites. The complete genome of the honey bee sequenced and variation among the genomic within species has been surveyed [38-40].
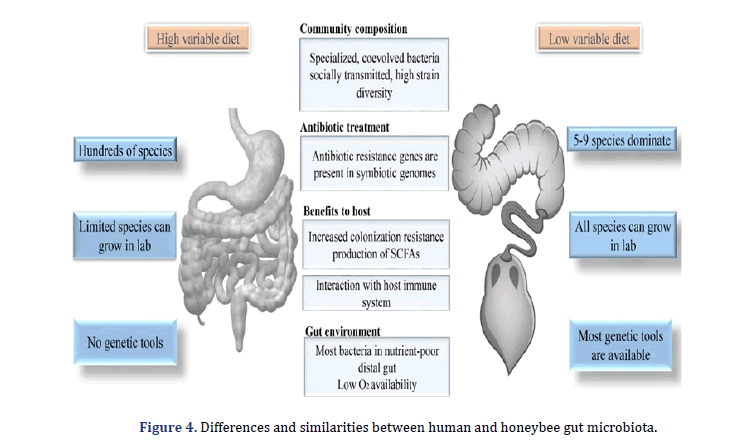
Apis mellifera’s microbiota is a major contributor among several social animals transmitted through direct contact with hive mates during social interaction [41-43]. The gut communities show high ecological resilience; despite the environmental changes, a distinctive organism group is maintained both within and between individuals [44,45]. The distinctive taxonomic makeup of the microbiota of social bees, together with their essential biochemical host contributions, suggests a highly functional, coevolved correlation between Apis mellifera and microbes [46,47]. In the end, the domesticated position of A.mellifera allows them a readily available system for microbiome research; that’s the reason why the gut ecosystem of A.mellifera plays a role as novel ecological facts with several possible applications [48,49] contributed to the first review of applicable methods for the culture and characterization of A. mellifera gut microbial communities. Recently, Zheng et al. Outlined the honeybee characteristic that makes an important experimental system for studies on gut microbiome via discussing established protocols and highlighting the comparison between the human hut and honeybee microbiotas [50]. Based on previously discussed strategies for studying the bee microbiota, our review emphasizes the latest developments and suggests a new idea in this field. The information summarized here can also be appropriate for the investigation of other insects with similarly developed microbiota, particularly within Hymenoptera.
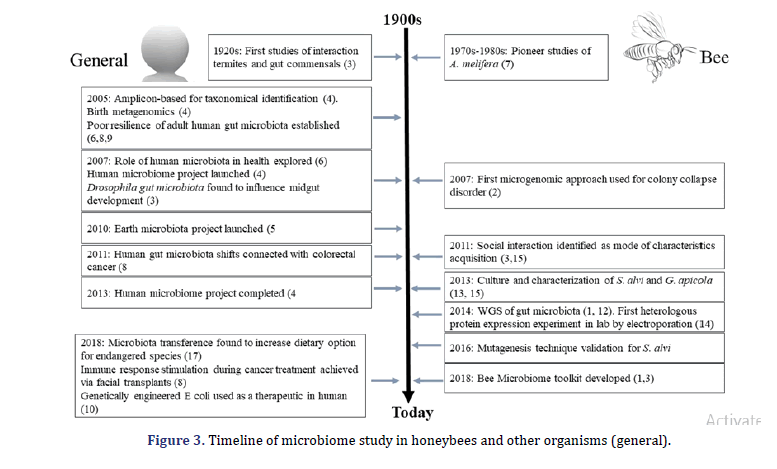
Microbial complex communities are present in almost every place on the human body. Still, the microbes are related to the Gastrointestinal (GI) tract, home to a wide range of microbes in several animals, are of specific interest to their various impact on the host’s health. The gut microbiota of humans bolsters potential anti-pathogen assists in food digestion and regulates the immune system. Sequencing analysis and sequencing have been for correlation identification between the diversity of disease and microbiota composition. Still, the experimental methods are crucial to getting through address cause, correlation, and effect relationship. As a practical and ethical study constraint on human experiments, the best model organism systems are important for experimental research of gut microbiota. In this review, we describe the honeybee gut microbiota as a model system that provides an experimentally tractable and offers several parallels to humans’ gut microbiota of humans (Figure 3).
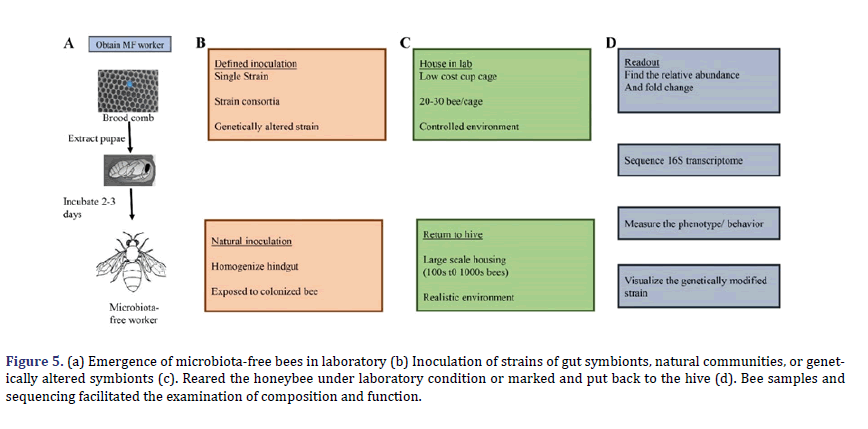
In (Figure 4), hive frames with capped brood (mature pupae) are separated from hives and transferred to the laboratory to create gnotobiotic bees. Pupae with eye pigmentation but incapable of moving are removed and kept in sterile dishes. Pupae are placed under hygienic conditions, then within three days, microbiota-free worker bees will emerge. Alternatively, worker bees with free microbiota are also reared in the laboratory by manually larval rearing, through this strategy needs more precise experimental infrastructure and less robust bees yield. The microbiota-free bees can be injected orally with whole communities or specific strains of bacteria to study the critical functions of gut microbiota and impact bee health and the mechanisms that microbes with one another and their host.
Effects on endocrine signalling and behaviour
A study associated with microbiota-free to conventional bee workers has revealed that the gut microbiota is needed for weight gain, but ileum and midgut microbiota-free workers are not more substantial than usual bees. The weight gain effect is related to shifts in gene expression, endocrine signaling, and modulation in Drosophila insulin- like/insulin signaling, also increased the vitellogenin level (nutritional modulator in honeybee). Kesnervoa et al. demonstrated that gram-positive bacteria (Bifidobacterium asteroides) stimulate the juvenile hormone derivatives and prostaglandins production is known to influence bee development [10,51].
Schwarz et al. discovered the down regulation expression of vitellogenin in gnotobiotic worker bees and under hive condition, mono inoculated with S.alvi and subsequent infection with Lotmaria passim (trypanosomatid parasite). As vitellogenin modulates the social behavior development in honey bees, these observations recommends an essential role of gut microbiota impacting the social behaviour of bee [52,53].
Many experiments have been conducted on the possible relation between behavior and gut microbiota of honeybees. Gut microbes may influence the host behavior by altering the biogenic amines level, such as dopamine, serotonin, and octopamine. Levels of biogenic amines in the brains of honeybee workers differ seasonally, and concentrations of amines increase in summer due to higher foraging activity [54]. Concentrations of amines in microbiota bees (newly emerged bees) are higher than the brains of adult bees (Conventional bees). Newly emerged bees and adult bees behave and respond differently; the sucrose of response of adult bees more readily and feeding more, which is regularly observed in insulin signaling. These findings provide strong evidence that hormonal signaling and host behavior are altered by gut microbes [55].
Effect on immune system
The microorganism group in symbiotic relations with host animals can be essential for host health. Particularly, insects severely harbor valuable gut microbiota, beneficial in disease resistance and food management [56]. The gut microbiota of A. mellifera comprises nearly nine species of transmitted bacteria that have an evolutionary link with their host [57,58]. Gut microbiota can modulate the host’s immune function, indirectly influencing host fitness and other microbes. Colonization by a single S.alvi or conventional microbiota resulted in the up-regulation of the hymenoptaecin and the antimicrobial peptides apidaecin in gut epithelial cells [59]. Frischella perrara (bacterium) plays a dramatic role in immune response in several honeybee species, colonising the honeybee’s pylorus in which midgut passes through the ileum. Colonization by bacterium triggers the formation of the ‘scab’ phenotypically looks like the dark ring around the gut, the formation of a dark ring caused by the melanization of an innate immune system in honeybee. However, F. perrara (a bacterium) interacts with the honey bee’s immune system [47].
The neurological effect of gut microbiota
The relation between neurophysiology, behavior, and gut microbes of hosts has exponentially grown in the last few years. Most research has focused on model organisms despite their broad significance for human health. For several reasons, the honey bee is an excellent model for studying bacterial symbionts’ neurological impact [60]. In the honeybee gut, a bacterial population comprised many sequence discrete populations SDPs, considered species of bacteria (Figure 5). Each bee is consisting a wide range of strain diversity. In each bee, distinctive strains combination represents that gut microbiota’s function differs in bees even in the same hive. Unique behavioral strains characterized through the division of labor present in the same hive and showed differences in gut microbiota structure and composition [61,62]. The behavior of worker bees is modulated by gut microbiota by increasing the level of sugar intake the same as by changing insulin sensitivity. Bifidobacterium asteroides induce prostaglandins and Juvenile hormone III in the host gut, which can be involved in brain-gut communication [63,64]. The research on the neurophysiological impact of gut microbes is in the initial stage, but the honeybee is a significant pollinator for securing food production. It could play an essential contribution to maintaining the hive health.
Figure 5. (a) Emergence of microbiota-free bees in laboratory (b) Inoculation of strains of gut symbionts, natural communities, or genetically altered symbionts (c). Reared the honeybee under laboratory condition or marked and put back to the hive (d). Bee samples and sequencing facilitated the examination of composition and function.
Perturbation of the native microbiota
Perturbation of established and normal gut community, using disruptors and antibiotics, provides more microbiota function information. Raymann et al. examined the treatment effects with tetracycline, broad range spectrum, gut microbiota composition of the honeybee, and host fitness [65-67]. Bees treated by antibiotics showed modified relative diversity and abundance of core microbial taxa, elevated non-core taxa abundance, and increased mortality rate and lower survival rate whenever exposure to the Serratia marcescens kz11 (opportunistic pathogen). Likewise, none of the dominant microbiota members were eliminated despite antibiotic treatment. Gut microbiota was harmful even opportunistic pathogens were not present; under laboratory conditions, adult bees showed a relatively higher mortality rate than microbiota free-bees after being treated with the antibiotic [58]. Li et al. revealed that disturbance in the microbiota of worker bees with antibiotics decreased the immune response and increased the susceptibility to Nosema ceranae (microsporidian parasite) that invasion by the midgut epithelium [50,68].
The structural difference of honeybee gut microbiota in summer and winter
Adult bees contain specialized and relatively less complex gut microbiota. In comparison, the composition of the gut communities has differences in the summer and winter seasons. The amount and type of nutrients (i.e., nectar and pollen) present throughout the foraging period can profoundly affect the metabolic activity and gut microbiota composition [69-71]. Likewise, different dietary habitats and lifespan variations of worker bees throughout the winter and summer may affect gut microbiota’s composition [70,72,73] . The surprising finding in the winter bees is that gut microbiota lives longer in the foraging season, which is critical for the colony’s survival and health. In cold weather, most colony losses occur due to limited resources [74-76].
The gut microbiota of honey bees and human
Although bee health is a significant reason for investigating the gut microbiota of honeybees, the main advantages of this system are that there are many parallels to human gut microbiota.
Evolutionary and specificity to hosts
The gut microbiota community of both humans and honey bees has the same environment as the host gut. Gut bacteria in humans and honeybees are likely to be precisely adapted to the habitats, as they coexist across millions of years with their hosts [6,77,78]. The most abundant five bacterial species linked with the guts of Meliponini (stingless bees) corbiculate (family Apidae), Bombus (bumblebees), Apis (Asian honey bees) most possibly descend from the community exist in the ancestor of bees, with succeeding strain gain, losses, the divergence of taxa making the gut communities found [38,79].
Transmission pattern through social interaction
Both gut bacteria and bee are mainly transmitted; in honeybees, the evolutionary study of bacterial strains showed throughout corbiculate bee hosts recommend that their performance is associated with the change to social lifestyles [80-83]. Bees’ core microbiota is not present in the wasps or solitary bees; neither was isolated from the environment. In comparison, gut communities of many invertebrates have unpredictable compositions regulated via bacteria from the environmental source [84-86].
Strain variation of bacterial species
Although the gut microbiota of bees possesses bacterial species in a limited number, each species shows extensive variation in strain, which is the same as in the microbiota of the human gut. Next-generation sequencing of the gene (single cope protein-coding) exhibited high G. apicola and S. alvi strain diversity in honey bee guts [87,88]. Both species have large genetic pools of genes that do not exist in all strains. For instance, some accessory genes in the strains of G. apicola are contributed to carbohydrate metabolism; some strains are monosaccharides (toxic to host) and gene encoding for utilization of pollen cell walls components [88-90]. Several strains have distinguished assortments of T6SS, Type VI Secretion System-associated with antitoxin and toxin genes, which may affect which strains combination allows single host co-colonization. Furthermore, few strains of Apibacter (interbacterial antagonism by Bacteroidetes species) encoded similar to Type VI Secretion systems used in the human gut [16,91,92].
Negative effect on host health
The communities of the gut microbiome harm the health of the host. In humans, dysbacteriosis or abnormal function or composition of the microbiota is linked with several diseases and causes poor diet, antibiotic treatment, and other disturbances [93-100]. For example, disturbance of the gut microbiome due to antibiotics treatment decreases the resistance to infection by Clostridium difficile infections in humans. In the same way, disruption in the gut microbiota of honey bees due to chemicals or antibiotic treatment increases the susceptibility to S. marcescens infections.
Role in fermentation and short-chain fatty acid production
The bee gut microbiota is located in the distal gut in other animals and humans. It involved fermentation and digestion of carbohydrate polymers obtained from the plant’s cell walls. The function of bee guts microbiota differences with the gut microbiota of the other insects. For example, in the fruit fly Drosophila melanogaster, gut microbiota colonized in the midgut and not concerned with digesting the plant cell wall component; however, it is vital developmental and immune signaling [101-106]. Even herbivores larvae of lepidopteran, which eat just plant material, seem not to depend upon the gut microbiome for nutrition and digestion. The availability of oxygen in the gut can affect the colonization pattern and influence the mutual relationship between gut microbiota. The guts of herbivorous insects are almost anoxic compared to D. melanogaster, controlled by aerobes and contains oxygen. Anoxia is maintained in the honey bee ileum through the S. alvi respiration; S. alvi is a bacterium linked with the ileum wall, which is driven by acetate, the abundant short-chain fatty acid in the gut [107-112].
History and exposure to antibiotic
Long-term use of antibiotics may have influenced the diversity in humans’ gut communities and caused a high level of resistance factors. Similarly, antibiotic exposure has impacted the bee gut communities, especially in those countries where most beekeepers have applied antibiotics to prevent or control the foulbrood (a larval bacterial disease) [112-115]. The use of antibiotics resulted in resistance in gut bacteria and isolated the bee from those countries in which beekeepers were not allowed to practice antibiotics in beekeeping. In both honey bee and human gut communities, the resistance factors have replaced the community members by horizontal transfer. Additionally, exposure to antibiotics directly influences the diversity and size of bee gut communities [116-120].
What is the functional role of the gut microbiome in different weather and colony health?
Colony losses and mortality rates depend upon the weather seasons, including spring, summer, autumn, and winter. The gut microbiota community showed the difference in different weather conditions in adult bees. Moreover, winter bees showed limited information about gut microbiota [121-125]. During the foraging season in winter, gut microbiota lives longer than adult bees which are severe threats to the colony’s survival. The mortality rate of bees is higher in winter weather than in other weather [126- 128]. In the future, further investigation is required to understand the life span and regulation of gut microbiota in adult bees during the winter season.
Discussion and Conclusion
This review illustrated the various advantages of model organisms for microbiota research in this review. Simple model organisms can provide mutual purposes to study the cellular and molecular mechanisms that support host-microbiome interaction, which is already identified in model animals and humans. These benefits include intrinsic characteristics, the ability to colonize to study microbes and host interaction, and the rearing of bees economically. For years, honeybee research has had a knowledge base in multiple fields: behavioural, experimental protocols, ecological, developmental, genomics, and physiological information. This research’s motivation is that the gut microbiota of honey bees provides evidence for its essential functions related to preventing colony losses, preserving pollinators, and bee health.
Moreover, the gut microbiota of the bee and humans has several similarities. In this concern, a major reason for using the simple model organisms is described as the interactions of host and microbiota, which are possibly related to humans also. The meaningful purpose of this review is that many appropriate simple model species can contribute significantly to our understanding of animals and microbiome interactions. In the coming years, it is expected that leading findings on basics of host-microbiome interaction=ions from research on the traditional animal models, controlled by tools and excellent resources commanded by these species. Research on the behavioural phenotypes of gut microbes symbionts has consequences across medical and biological disciplines. In future research, to increase the value of bacterial symbionts’ role in the social brain evolution, The first encouraging investigation has recommended that homologous brain and gut microbiota interactions in increase and mammals may exist, indicating a profound evolutionary origin brain axis and gut microbiota. Establishing the function of gut microbiota in behavior, cognition and prebiotic dietary supplementary as a method to regulate the behavioral characteristics of animals as the strategic significance has the great potential to build up a distinct aspect on how honey bees, as well as other insects, will be handled in the future. Another critical question is how gene expression changes in the brain interact with the brain’s neuron connection to affect behavior? In the future, further studies on the roles of miRNA, alternative spicing, and epigenetic in regulating bee behavior and gene expression, which and what degree in protein-coding sequence and gene expression enables changes in behavior? The most important question is how novel and conserved behavioral genes relate to evolutionary and mechanistic contexts? What is the relationship between behavioral and neurogenic as a general phenomenon? A burgeoning review of literature has related differences in brain expression to regulate the behavioral changes in many species, but more work is needed to explore the role of gut microbiota in honey bees and other simple model organisms.
Authors’ Contribution
Muhammad Fahad Raza and Muhammad Asad substantially contributed to the conception and design of the article and interpreted the relevant literature. Muhammad Fahad Raza, Ammar Raza and Bilal Atta drafted the article or revised it critically for important intellectual content.
REFERENCES
- Christian N, Whitaker BK, Clay K. Microbiomes: Unifying animal and plant systems through the lens of community ecology theory. Front Microbiol 2015; 6: 869.
[Crossref] [Google scholar] [Pubmed]
- Bahrndorff S, Alemu T, Alemneh T, Nielsen JL. The microbiome of animals: Implications for conservation biology. Int J Genomics 2016; 5304028
[Crossref] [Google scholar] [Pubmed]
- Bang C, Dagan T, Deines P, Dubilier N, Duschl WJ, Fraune S, et al. Metaorganisms in extreme environments: Do microbes play a role in organismal adaptation? Zoology (Jena) 2018; 127: 1-19.
[Crossref] [Google scholar] [Pubmed]
- Kostic AD, Howitt MR, Garrett WS. Exploring host–microbiota interactions in animal models and humans. Genes Dev 2013; 27 (7): 701-18.
[Crossref] [Google scholar] [Pubmed]
- Douglas AE. Simple animal models for microbiome research. Nat Rev Microbiol 2019; 17: 764-75.
[Crossref] [Google scholar] [Pubmed]
- Engel P, Moran NA. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol Rev 2013; 37 (5): 699-735.
[Crossref] [Google scholar] [Pubmed]
- Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature 2017; 548: 43-51.
- Anjum SI, AH Shah, M Aurongzeb, J Kori, MK Azim, MJ Ansari, et al. Characterization of gut bacterial flora of apis mellifera from north-west pakistan. Saudi J of Biol Sci 2018; 25 (2): 388-92.
- Romero S, Nastasa A, Chapman A, Kwong WK, Foster LJ. The honey bee gut microbiota: Strategies for study and characterization. Insect Mol Biol 2019; 28 (4): 455-72.
[Crossref] [Google scholar] [Pubmed]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. PNAS 2013; 110: 3229-36.
- Vanner SJ, Greenwood-Van Meerveld B, Mawe GM, Shea-Donohue T, Verdu EF, Wood J, et al. Fundamentals of neurogastroenterology: Basic science. Gastroenterology 2016; 150: 1280-91.
[Crossref] [Google scholar] [Pubmed]
- Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive Gut Microbiota of Honey Bees Assessed Using Deep Sampling from Individual Worker Bees. PLoS One 2012; 7: e36393.
- Audisio MC.Gram-positive bacteria with probiotic potential for the apis mellifera l. Honey bee: The experience in the northwest of argentina. Probiotics Antimicrob Proteins 2017; 9 (1): 22-31.
[Crossref] [Google scholar] [Pubmed]
- Koyle ML, Veloz M, Judd AM, Wong ACN, Newell PD, Douglas AE, et al. Chaston, 2016. Rearing the fruit fly drosophila melanogaster under axenic and gnotobiotic conditions. J Vis Exp 2016; 113: e54219.
[Crossref] [Google scholar] [Pubmed]
- Mushegian AA, Walser J-C, Sullam KE, Ebert D. The microbiota of diapause: How host-microbe associations are formed after dormancy in an aquatic crustacean. J Anim Ecol 2018. 87 (2): 400-13.
- Samuel BS, Rowedder H, Braendle C, Félix MA, Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc Natl Acad Sci U S A 2016; 113 (27): E3941-9.
[Crossref] [Google scholar] [Pubmed]
- Leulier F, MacNeil LT, Lee WJ, Rawls JF, Cani PD, Schwarzer M, et al. Integrative physiology: At the crossroads of nutrition, microbiota, animal physiology, and human health. Cell Metab 2017; 25 (3): 522-34.
[Crossref] [Google scholar] [Pubmed]
- Flavel MR, Mechler A, Shahmiri M, Mathews ER, Franks AE, Chen W, et al. Growth of caenorhabditis elegans in defined media is dependent on presence of particulate matter. G3 (Bethesda) 2018; 8 (2): 567-75.
[Crossref] [Google scholar] [Pubmed]
- Davis RH. The microbial models of molecular biology: From genes to genomes. Oxford University Press 2003;
- Belcaid M, Casaburi G, McAnulty SJ, Schmidbaur H, Suria AM, Moriano-Gutierrez S, et al. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc Natl Acad Sci U S A 2019; 116 (8): 3030-35.
[Crossref] [Google scholar] [Pubmed]
- Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 2014; 111: 11509-14.
[Crossref] [Google scholar] [Pubmed]
- Racz PI, Wildwater M, Rooseboom M, Kerkhof E, Pieters R, Yebra-Pimentel ES, et al. Application of caenorhabditis elegans (nematode) and danio rerio embryo (zebrafish) as model systems to screen for developmental and reproductive toxicity of piperazine compounds. Toxicol In Vitro 2017; 44: 11-16.
[Crossref] [Google scholar] [Pubmed]
- Zhang Y, Lu H, Bargmann C. Pathogenic bacteria induce aversive olfactory learning in caenorhabditis elegans. Nature 2005;438 (7065): 179-84.
[Crossref] [Google scholar] [Pubmed]
- Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc 2008; 3 (12):1862-75.
- Schulenburg H, Félix MA. The natural biotic environment of caenorhabditis elegans. Genetics 2017. 206 (1): 55-86;
- Zhang F, Berg M, Dierking K, Félix MA, Shapira M, Samuel BS, et al. Caenorhabditis elegans as a model for microbiome research. Front Microbiol 2017; 8: 485.
[Crossref] [Google scholar] [Pubmed]
- Lemaitre B, Miguel-Aliaga I. The digestive tract of drosophila melanogaster. Annu Rev Genet 2013; 47: 377-404.
[Crossref] [Google scholar] [Pubmed]
- Miguel-Aliaga I, Jasper H, Lemaitre B. Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 2018; 210 (2): 357-96.
- Pais IS, Valente RS, Sporniak M, Teixeira L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol 2018; 16 (7): e2005710.
- Medrzycki P, Giffard H, Aupinel P, Belzunces LP, Chauzat MP, Claßen C, et al. Standard methods for toxicology research in Apis mellifera. Journal of Apicultural Research 2013; 52 (4): 1-60. [Crossref]
[Google scholar][PubMed]
- Paudel Y, Mackereth R, Hanley R, Qin W. Honey Bees (Apis mellifera L.) and Pollination Issues: Current status, impacts and potential drivers of decline. Journal of Agricultural Science 2015; 7 (6): 93.
- Saunders ME, Peisley RK, Rader R, Luck GW. Pollinators, pests, and predators: Recognizing ecological trade-offs in agroecosystems. Ambio 2016; 45: 4-14.
[Crossref] [Google scholar] [Pubmed]
- Robinson GE, Page RE, Strambi C, Strambi A. Hormonal and genetic control of behavioral integration in honey bee colonies. Science 1989; 246 (4926): 109-12.
[Crossref] [Google scholar] [Pubmed]
- Jones N, Salles JM, Settele J, Vaissière BE. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics 2009; 68 (3): 810-21.
- Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol 2016; 14: 374-84.
[Crossref] [Google scholar] [Pubmed]
- Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, et al. Dynamic microbiome evolution in social bees. Sci Adv 2017; 3 (3): e1600513.
[Crossref] [Google scholar] [Pubmed]
- Jones JC, Fruciano C, Marchant J, Hildebrand F, Forslund S, Bork P, et al. The gut microbiome is associated with behavioural task in honey bees. Insectes Soc 2018; 65 (3): 419-29.
[Crossref] [Google scholar] [Pubmed]
- Raymann K, Moran NA. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci 2018; 26: 97-104.
[Crossref] [Google scholar] [Pubmed]
- Easton-Calabria A, Demary KC, Oner JN. Beyond pollination: Honey bees (apis mellifera) as zootherapy keystone species. Frontiers in Ecol and Evolu 2019; 6: 161.
- Tauber JP, Collins WR, Schwarz RS, Chen Y, Grubbs K, Huang Q, et al. Natural product medicines for honey bees: Perspective and protocols. Insects 2019; 10 (10): 356.
- Kapheim KM, Rao VD, Yeoman CJ, Wilson BA, White BA, Goldenfeld N, et al. Caste-specific differences in hindgut microbial communities of honey bees (apis mellifera). PLoS One 2015;10: e0123911
[Crossref] [Google scholar] [Pubmed]
- Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, et al. The bee microbiome: Impact on bee health and model for evolution and ecology of host-microbe interactions. ASM Journals 2016; 7 (2): e02164-02115.
[Crossref] [Google scholar] [Pubmed]
- Steele MI, Kwong WK, Whiteley M, Moran NA. Diversification of type vi secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 2017; 8 (6): e01630-17.
[Crossref] [Google scholar] [Pubmed]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489 (7415): 220-30.
[Crossref] [Google scholar] [Pubmed]
- Shade A, Peter H, Allison SD, Baho D, Berga M, Bürgmann H, et al. 2012. Fundamentals of microbial community resistance and resilience. Front Microbiol 2012; 3: 417.
[Crossref] [Google scholar] [Pubmed]
- Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 2011; 20 (3): 619-28.
- Engel P, Bartlett KD, Moran NA. The bacterium frischella perrara causes scab formation in the gut of its honeybee host. mBio 2015; 6 (3): e00193-15.
[Crossref] [Google scholar] [Pubmed]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature 2011; 474 (7351): 327-36.
[Crossref] [Google scholar] [Pubmed]
- Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of Acquisition of the Gut Microbiota of the Honey Bee Apis mellifera. Appl Environ Microbiol 2014; 80 (23): 7378-87.
- Zheng H, Steele MI, Leonard SP, Motta EV, Moran NA. Honey bees as models for gut microbiota research. Nature 2018; 47: 317-25.
- Goblirsch M, Huang ZY, Spivak M. Physiological and behavioral changes in honey bees (apis mellifera) induced by nosema ceranae infection. PLoS One 2013; 8: e58165.
[Crossref] [Google scholar] [Pubmed]
- Ihle KE, Baker NA, Amdam GV. Insulin-like peptide response to nutritional input in honey bee workers. J Insect Physiol 2014; 69: 49-55.
[Crossref] [Google scholar] [Pubmed]
- Schwarz RS, Moran NA, Evans JD. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc Natl Acad Sci U S A 2016; 113 (33): 9345-50.
[Crossref] [Google scholar] [Pubmed]
- Chen YL, Hung YS, Yang EC. Biogenic amine levels change in the brains of stressed honeybees. Arch Insect Biochem Physiol 2008; 68 (4): 241-50.
[Crossref] [Google scholar] [Pubmed]
- Bordier C, Klein S, Conte YL, Barron AB, Alaux C. Stress decreases pollen foraging performance in honeybees. J Exp Biol 2018; 221 (4): jeb171470.
[Crossref] [Google scholar] [Pubmed]
- Kwong WK, Moran NA, Evolution of host specialization in gut microbes: The bee gut as a model. Gut Microbes 2015. 6 (3): 214-20.
[Crossref] [Google scholar] [Pubmed]
- Li G, Zhao H, Liu Z, Wang H, Xu B, Guo X. The wisdom of honeybee defenses against environmental stresses. Front Microbiol 2018; 9: 722.
[Crossref] [Google scholar] [Pubmed]
- Rouzé R, Moné A, Delbac F, Belzunces L, Blot N. The honeybee gut microbiota is altered after chronic exposure to different families of insecticides and infection by nosema ceranae. Microbes Environ 2019; 34 (3): 226-33.
[Crossref] [Google scholar] [Pubmed]
- Shanahan F. The host–microbe interface within the gut. Best Pract Res Clin Gastroenterol 2002; 16 (6): 915-31.
[Crossref] [Google scholar] [Pubmed]
- Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, et al. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS One 2012; 7 (3): e33188.
[Crossref] [Google scholar] [Pubmed]
- Jones JC, Fruciano C, Hildebrand F, Al Toufalilia H, Balfour NJ, Bork P, et al. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol Evol 2017; 8 (1): 441-51.
[Crossref] [Google scholar] [Pubmed]
- Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Description of snodgrassella alvi gen. Nov., sp. Nov., a member of the family neisseriaceae of the betaproteobacteria, and gilliamella apicola gen. Nov., sp. Nov., a member of orbaceae fam. Nov, orbales ord. Nov, a sister taxon to the order ‘enterobacteriales’ of the gammaproteobacteria. Int J Syst Evol Microbiol 2013; 63 (6): 2008-18.
[Crossref] [Google scholar] [Pubmed]
- Ellegaard KM, Brochet S, Bonilla‐Rosso G, Emery O, Glover N, Hadadi N, et al. Genomic changes underlying host specialization in the bee gut symbiont lactobacillus firm5. Microbiology 2019; 28 (9): 2224-37.
- Kešnerová L, Emery O, Troilo M, Liberti J, Erkosar B, Engel P. Gut microbiota structure differs between honeybees in winter and summer. ISME J 2020; 14: 801-14.
[Crossref] [Google scholar] [Pubmed]
- Liberti J, Engel P. The gut microbiota—brain axis of insects. Curr Opin Insect Sci 2020; 39: 6-13.
[Google scholar] [Pubmed]
- Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol 2017; 15 (3): e2001861.
[Crossref] [Google scholar] [Pubmed]
- Kim WH, Lillehoj HS. Immunity, immunomodulation, and antibiotic alternatives to maximize the genetic potential of poultry for growth and disease response. Animal Feed Sci and Techn, 2019; 250: 41-50.
- Klancic T, Reimer AR. Gut microbiota and obesity: Impact of antibiotics and prebiotics and potential for musculoskeletal health. J Sport Health Sci 2020; 9 (2): 110-18.
[Crossref] [Google scholar] [Pubmed]
- Panek J, Paris L, Roriz D, Mone A, Dubuffet A, Delbac F, Diogon M et al.Impact of the microsporidian Nosema ceranae on the gut epithelium renewal of the honeybee. Apis mellifera. J Invertebr Pathol 2018; 159: 121-28.
- Remolina SC, Hafez DM, Robinson GE, Hughes KA. Senescence in the worker honey bee apis mellifera. J Insect Physiol 2007; 53 (10): 1027-33.
[Crossref] [Google scholar] [Pubmed]
- Brodschneider R, Crailsheim K, Nutrition and health in honey bees. Nutrition et santé des abeilles 2010; 41: 278-94.
- Lau P, Bryant V, Ellis JD, Huang ZY, Sullivan J, Schmehl DR, et al. Seasonal variation of pollen collected by honey bees (apis mellifera) in developed areas across four regions in the united states. PLoS One 2019; 14: e0217294.
[Crossref] [Google scholar] [Pubmed]
- Di Pasquale G, Salignon M, Le Conte Y, Belzunces LP, Decourtye A, Kretzschmar A, et al. Influence of pollen nutrition on honey bee health: Do pollen quality and diversity matter? PLoS One 2013; 8 (8): e72016.
[Crossref] [Google scholar] [Pubmed]
- Anderson KE, VA Ricigliano, BM Mott, DC Copeland, AS Floyd, Maes P. The queen’s gut refines with age: Longevity phenotypes in a social insect model. Microbiome 2018; 6: 1-16.
[Crossref] [Google scholar] [Pubmed]
- Genersch E, Von Der Ohe W, Kaatz H, Schroeder A, Otten C, Büchler R, et al. The german bee monitoring project: A long term study to understand periodically high winter losses of honey bee colonies. Nature 2010; 41: 332-52.
- Braga AR, Gomes DG, Rogers R, Hassler EE, Freitas BM, Cazier JA. A method for mining combined data from in-hive sensors, weather and apiary inspections to forecast the health status of honey bee colonies. Comput and Elect in Agriculture 2020; 169: 105161.
- Kešnerová L, Emery O, Troilo M, Liberti J, Erkosar B, Engel P. Gut microbiota structure differs between honeybees in winter and summer. ISME J 2020; 14: 801-14.
[Crossref] [Google scholar] [Pubmed]
- Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, et al. Dynamic microbiome evolution in social bees. Sci Adv 2017; 3 (3): e1600513.
[Crossref] [Google scholar] [Pubmed]
- Segers FHID, Kaltenpoth M, Foitzik S. Abdominal microbial communities in ants depend on colony membership rather than caste and are linked to colony productivity. Ecology and Evolution 2019; 9: 13450-67.
- Leonhardt SD, Kaltenpoth M. Microbial communities of three sympatric australian stingless bee species. PLoS One 2014. 9 (8): e105718.
[Crossref] [Google scholar] [Pubmed]
- Archie EA, Tung J. Social behavior and the microbiome. Current Opin in Behavi Scie 2015; 6: 28-34.
[Google scholar] [Pubmed]
- Dinan TG, Stilling RM, Stanton C, F Cryan J. Collective unconscious: How gut microbes shape human behavior. J of Psychi Rese 2015. 63: 1-9.
- Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, et al. Cospeciation of gut microbiota with hominids. Science 2016; 353 (6297): 380-82.
- Sarkar A, Harty S, Lehto SM, Moeller AH, Dinan TG, Dunbar RI, et al. The microbiome in psychology and cognitive neuroscience. Trends Cogn Sci 2018; 22 (7): 611-36.
[Crossref] [Google scholar] [Pubmed]
- Emery NJ, Seed AM, von Bayern AMP, Clayton NS. Cognitive adaptations of social bonding in birds. Philos. Trans R Soc Lond B Biol Sci 2007; 362 (1480): 489-505.
[Crossref] [Google scholar] [Pubmed]
- McFrederick QS, Thomas JM, Neff JL, Vuong HQ, Russell KA, Hale AR, et al. Flowers and Wild Megachilid Bees Share Microbes. Microb Ecol 2017; 73 (1): 188-200.
- Keller A, Brandel A, Becker MC, Balles R, Abdelmohsen UR, Ankenbrand MJ, et al. Wild bees and their nests host paenibacillus bacteria with functional potential of avail. Microbiome 2018; 6: 229.
[Crossref] [Google scholar] [Pubmed]
- Kwong WK, Moran NA. Evolution of host specialization in gut microbes: The bee gut as a model. Gut Microbes 2015; 6 (3): 214-220.
[Crossref] [Google scholar] [Pubmed]
- Steele MI, Kwong WK, Whiteley M, Moran NA. Diversification of type vi secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 2017; 8 (6): e01630-17.
[Crossref] [Google scholar] [Pubmed]
- Engel P, Stepanauskas R, Moran NA. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet 2014; 10 (9): e1004596.
[Crossref] [Google scholar] [Pubmed]
- Ellegaard KM, Engel P. Genomic diversity landscape of the honey bee gut microbiota. Nature Communications 2019; 10: 1-13.
- Kwong WK, Steele MI, Moran NA. Genome sequences of apibacter spp., gut symbionts of asian honey bees. Genome Biol Evol 2018; 10 (4): 1174-79.
[Crossref] [Google scholar] [Pubmed]
- Navarro-Garcia F, Ruiz-Perez F, Cataldi A, Larzábal M. Type VI Secretion System in Pathogenic Escherichia coli: Structure, Role in Virulence, and Acquisition. Front Microbiol 2019; 10: 1965.
- Maruščáková IC, Schusterová P, Bielik B, Toporčák J, Bíliková K, Mudroňová D. Effect of Application of Probiotic Pollen Suspension on Immune Response and Gut Microbiota of Honey Bees (Apis mellifera). Probiotics Antimicrob Proteins 2020; 12 (3): 929-36.
- Paxton RJ. A microbiome silver bullet for honey bees. Science 2020; 367 (6477): 504-06.
- Zheng H, Powell JE, Steele MI, Dietrich C, Moran NA. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. 2017; 114 (18): 4775-80.
- Cox CR, Gilmore MS. Native microbial colonization of drosophila melanogaster and its use as a model of enterococcus faecalis pathogenesis. Infect Immun 2007; 75 (4): 1565-76.
[Crossref] [Google scholar] [Pubmed]
- Tegtmeier D, Thompson CL, Schauer C, Brune A. Oxygen affects gut bacterial colonization and metabolic activities in a gnotobiotic cockroach model. Applied and Environmental Microbiology 2015; 82 (4): 1080-89.
- Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol 2017; 15 (3): e2001861.
[Crossref] [Google scholar] [Pubmed]
- Evans JD, Armstrong TN. Inhibition of the american foulbrood bacterium, paenibacillus larvae larvae, by bacteria isolated from honey bees. J of Apicul Rese 2005; 44 (4): 168-71.
- Daisley BA, Pitek AP, Chmiel JA, Al KF, Chernyshova AM, Faragalla KM, et al. Novel probiotic approach to counter paenibacillus larvae infection in honey bees. ISME J 2020; 14 (2): 476-91.
[Crossref] [Google scholar] [Pubmed]
- Peghaire E, Moné A, Delbac F, Debroas D, Chaucheyras-Durand F, El Alaoui H. A Pediococcus strain to rescue honeybees by decreasing Nosema ceranae- and pesticide-induced adverse effects. Pestic Biochem Physiol 2020; 163: 138-46.
- Alpatov WW. Biometrical studies on variation and races of the honey bee (apis mellifera L.). The Quarterly Review of Biology 1929; 4 (1): 1-58. [Crossref]
- Aron-Wisnewsky J, K Clément. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol 2016; 12: 169-81.
[Crossref] [Google scholar] [Pubmed]
- Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A 2007; 104 (3): 979-84.
[Crossref] [Google scholar] [Pubmed]
- Blyton MD, Soo RM,Whisson D, Marsh KJ, Pascoe J, Pla ML, et al. Faecal inoculations alter the gastrointestinal microbiome and allow dietary expansion in a wild specialist herbivore, the koala. Anim Microbiome 2019; 1: 1-18.
[Crossref] [Google scholar] [Pubmed]
- Bramoullé Y, Djebbari H, Fortin B. Identification of peer effects through social networks. Jour of Econo 2009; 150 (1): 41-55.
- Brock WA, Durlauf SN. Identification of binary choice models with social interactions. Jour of Econ 2007; 140: 52-75.
- Buchon N, Broderick NA, Lemaitre B. Gut homeostasis in a microbial world: Insights from drosophila melanogaster. Nat Rev Microbiol 2013; 11: 615-26.
[Crossref] [Google scholar] [Pubmed]
- Coté S. A social interaction model of the effects of emotion regulation on work strain. he Academy of Management Review 2005; 30 (3): 509-30.
- Graham BS. Identifying social interactions through conditional variance restrictions. Econometrica 2008; 76 (3): 643-60.
- Gross CL, Mackay D. Honeybees reduce fitness in the pioneer shrub melastoma affine (melastomataceae). Biological Conservation 1998; 86 (2): 169-78.
- Hartmann WR, Manchanda P, Nair H, Bothner M, P. Dodds, Godes D, et al. Modeling social interactions: Identification, empirical methods and policy implications. Marketing Letters 2008; 19: 287-04.
- Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019; 7: e7502.
[Crossref] [Google scholar] [Pubmed]
- Liu H, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 2008; 8: 1-10. [Crossref]
[Google scholar] [Pubmed]
- Liu X, Davis RL. Insect olfactory memory in time and space. Curr Opin Neurobiol 2006; 16 (6): 679-85.
[Crossref] [Google scholar] [Pubmed]
- Xia Y, Sun J. Hypothesis testing and statistical analysis of microbiome. Genes Dis 2017; 4 (3): 138-48.
[Crossref] [Google scholar] [Pubmed]
- Unger S, Stintzi A, Shah P, Mack D, O'Connor DL. Gut microbiota of the very-low-birth-weight infant. Pediatr Res 2015; 77 (1-2): 205-13.
[Crossref] [Google scholar] [Pubmed]
- Ursell LK, Haiser HJ, Van W, Treuren N, Garg L, Reddivari J, et al. The intestinal metabolome: An intersection between microbiota and host. Gastroenterology 2014; 146 (6): 1470-76.
[Crossref] [Google scholar] [Pubmed]
- Li W, Deng Y, Chu Q, Zhang P. Gut microbiome and cancer immunotherapy. Cancer Lett 2020; 447: 41-47.
- Suenami S, Nobu MK, Miyazaki R. Community analysis of gut microbiota in hornets, the largest eusocial wasps, vespa mandarinia and v. Simillima. Scientific Reports 2019; 9: 1-13.
- Miller KI. Social Bacteria in the Eusocial Bee: Investigating Host-Microbiome Interactions within the European Honey Bee (Apis mellifera). ProQuest Dissertations 2018; 10841004.
- Nagpal R, Kumar M, Yadav AK, Hemalatha R, Yadav H, Marotta F, et al. Gut microbiota in health and disease: an overview focused on metabolic inflammation. Benef Microbes 2016; 7 (2): 181-94.
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486: 207-14.
- Ntranos A, Casaccia P. The Microbiome-Gut-Behavior Axis: Crosstalk Between the Gut Microbiome and Oligodendrocytes Modulates Behavioral Responses. Neurotherapeutics 2018; 15 (1): 31-35.
- Obregon-Tito AJ, Tito RY, Metcalf J. Sankaranarayanan K, Clemente JC, Ursell LK, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nature Communications 2015; 6: 6505.
- Rosano GL, Ceccarelli EA. Recombinant protein expression in escherichia coli: Advances and challenges. Front. Microbiol 2014; 5: 172.
[Crossref] [Google scholar] [Pubmed]
- Selma MV, Espin JC, Tomas-Barberan FA. Interaction between phenolics and gut microbiota: Role in human health. J Agric Food Chem 2009; 57 (15): 6485-01.
[Crossref] [Google scholar] [Pubmed]
Copyright: © 2022 The Authors. This is an open access article under the terms of the Creative Commons Attribution NonCommercial ShareAlike 4.0 (https://creativecommons.org/licenses/by-nc-sa/4.0/). This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.